The laboratory research is constantly evolving, also depending on the people contributing to the several projects. At the moment, we are focusing on the following areas.
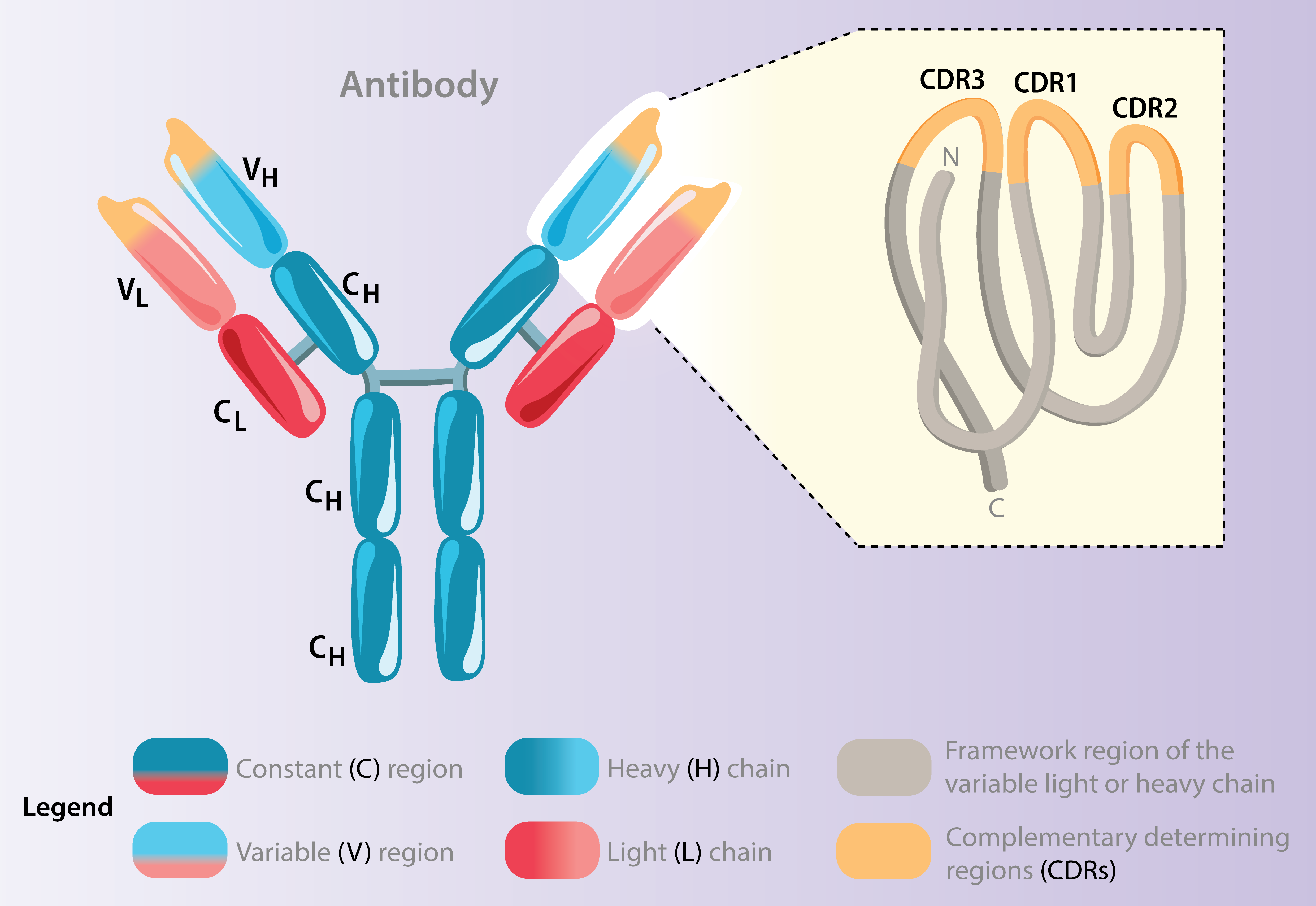
The antibody specificity highly depends on the complementarity determining regions (CDRs) of the heavy and light chain genes. We investigate the characteristics and mutations of these in many of our projects in order to determine e.g. clonal lineage tracing.
B cell responses in allergen immunotherapy
Allergen-specific immunotherapy (AIT) remains the only available treatment that targets the cause of IgE-mediated allergies. By repeatedly exposing allergic individuals to defined doses of allergen, AIT can significantly reduce symptoms and improve quality of life. It is particularly effective in treating seasonal allergies such as grass and tree pollen allergy.
AIT not only induces clinical tolerance but also reshapes both the cellular and humoral branches of the immune response.
We use AIT as a well-defined model of chronic antigen exposure to study how B cells evolve over time. This unique setting allows us to investigate key immunological processes, especially affinity maturation and B cell selection, in a controlled and longitudinal manner.
Our goal is to decipher the dynamics of allergen-specific B cell responses during AIT using cutting-edge approaches such as immunoglobulin repertoire sequencing and single-cell transcriptomics. By tracking these responses over several years, we aim to gain deeper insight into how chronic allergen exposure shapes the B cell repertoire and contributes to long-term immune modulation.
Organoids as a model system for allergy
So far, the underlying mechanisms of how B cells become activated during allergic responses are poorly understood. In this project we aim to utilize a tonsil tissue-on-chip model to analyze B cell activation in IgE-mediated allergy and to assess the underlying mechanisms on cellular and genetic level.
This cutting-edge method for culturing patient-derived tonsil organoids on a microfluidic chip provides a novel platform that replicates human tonsil tissue to study adaptive immune responses in vitro .
These organoids will be exposed to defined allergens like birch and grass pollen, including recombinant Bet v 1, Phl p 1, Phl p 5, and TLR agonists (Pam3CSK4 and flagellin) to drive B cell activation. Finally, single-cell multiomics (scRNA-seq and scATAC-seq) will be used to profile immune responses, B cell activation states, and IgE-related transcriptional programs.
Primary immunodeficiencies affecting B cell responses
Common Variable Immunodeficiency (CVID) is characterized by defective antibody production and impaired B cell differentiation. To better understand the origins of this dysfunction, we focus on both naïve and memory B cell compartments.
In our recent study (Friman et al., Cell Reports, 2023), we performed Ig light chain repertoire sequencing of naïve and CD27bright memory B cells (MBCs), revealing impaired peripheral selection and reduced somatic hypermutation in CVID patients, particularly those with autoimmune manifestations. These findings suggest a failure in the transition from naïve to germinal center–derived MBCs. Additionally, bulk RNA sequencing of in vitro–activated naïve B cells uncovered defective activation responses and insufficient induction of mismatch repair genes in CVID, indicating compromised affinity maturation.
In an ongoing follow-up study, we are now further examining both ex vivo and in vitro–activated naïve and CD27bright MBCs from CVID patients and healthy controls. Preliminary data show that even ex vivo isolated naïve B cells already exhibit transcriptional alterations in CVID. Upon in vitro activation, these disturbances intensify, particularly in germinal center–related pathways. We are currently validating these findings at the protein level to further understand how early defects in the B cell compartment contribute to CVID pathogenesis.
Building on these insights, we aim to extend our analyses to patient groups with defined germinal center defects such as individuals with severe combined immunodeficiency (SCID) to further dissect the developmental checkpoints that regulate human B cell function and tolerance.
B-cell receptor sequencing analysis in health and disease
B cell receptor (BCR) sequencing provides a powerful tool to explore how B cells develop, adapt, and differentiate in response to antigens. Our projects leverage this approach across multiple contexts; healthy individuals, patients undergoing allergen-specific immunotherapy (AIT), and experimental mouse models; to understand how B cell selection and memory formation are shaped in different immune environments.
In healthy individuals, we study the baseline structure and diversity of the BCR repertoire across B cell subsets with a particular focus on MBCs to map principles of clonal selection and tolerance. In parallel, in AIT-treated patients, we longitudinally track allergen-reactive B cell clones using high-throughput Ig sequencing (see section on AIT).
To mechanistically probe memory B cell differentiation and localization, we employ transgenic mouse models with spontaneous germinal centre formation to allow lineage tracing. Using this system, we can follow germinal center–experienced B cells over time and track their persistence, migration, and fate over time. Our ongoing work is indicating MBCs derived from germinal centers preferentially localizes to the marginal zone of the spleen, a finding that aligns with older histological results and suggests a specialized role for marginal zone–resident memory B cells in long-term immune surveillance.
Together, these studies aim to define the molecular and spatial logic of B cell memory formation in both human and murine systems.
